Clustered matrix approximations: Test 5
Rectangular matrix A, with dense non-diagonal block structure
Contents
Load data
clear % load processed NotreDame data load('NotreDame_actors_preprocessed')
Clustering with GRACLUS
% max cluster size maxClustSize = 12000; [m,n] = size(A); B = [sparse(m,m) A; A' sparse(n,n)]; % load partitioning result to suppress graclus outputs for publish purposes %[prt,obj] = recGraclus(B,maxClustSize); load test5var clear B % Reorder vertices rprt = prt(1:m); cprt = prt(m+1:end); [rind,rprtsz,rprtc] = reorderVertices(rprt); [cind,cprtsz,cprtc] = reorderVertices(cprt); % Visualize clustering structure of A spy(A(rind,cind))
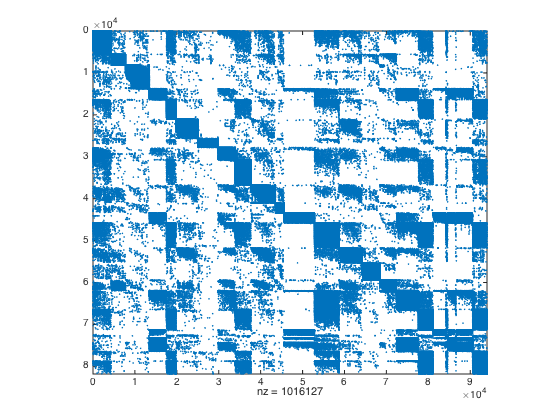
Determine and visualise location of dense blocks
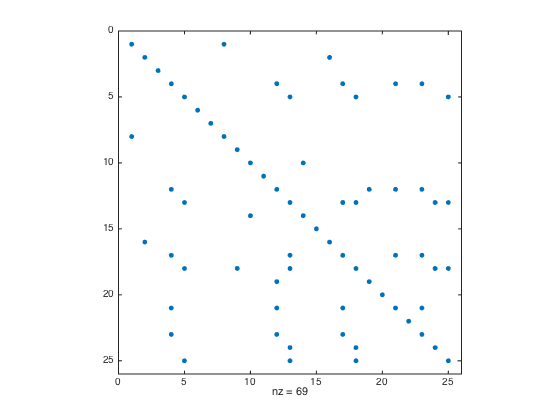
threshold = 0.15; [msk,~,totFrc] = determineDenseBlocks(A,rprtc,cprtc,threshold); clf spy(msk) nDenseBlocks = nnz(msk);
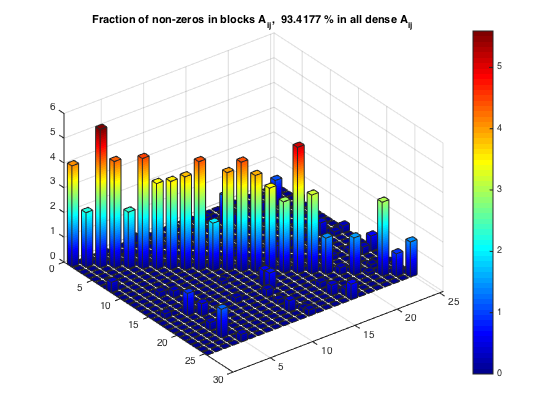
Fraction of non-zeros within diagonal blocks
[nnzBlks, nnzBlFr] = compClustNnzRect(A,rprtc,cprtc); bar3custom(msk.*nnzBlFr) title(['Fraction of non-zeros in blocks A_{ij}, ', num2str(totFrc), ... ' % in all dense A_{ij}']) % Relative error due to clustering: || A - A_dense || / || A || % where A_dense only contains non-zeros from dense blocks. relerr = compClustRelErrExt(A,rprtc,cprtc,msk); fprintf('Fraction of non-zeros in dense blocks : %8.2f %% \n', totFrc) fprintf('Relative error in a block diagonal approx.: %8.2f %% \n', relerr)
Fraction of non-zeros in dense blocks : 93.42 % Relative error in a block diagonal approx.: 25.66 %
Compute clustered matrix approximatons
% Specify rank k for approximations of dense blocks % Or use different ranks for different blocks k = 20; K = (ones(size(msk))*k).*msk; % Compute approximations for dense blocks of a rectangular matrix [U,S,V] = clustSvdsRect(A,K,rprtc,cprtc,'svds'); % if lansvd is available and prefered %[U,S,V] = clustSvdsRect(A,rprtc,cprtc,k,'lansvd'); % Specify parameters for clustered randomized algorithms p = 2; P = (ones(size(msk))*p).*msk; q = 2; Q = (ones(size(msk))*q).*msk; [Ur,Sr,Vr] = clustRandAlgRect(A,K,P,Q,rprtc,cprtc); SS = cell2mat(S); SSr = cell2mat(Sr);
Compute relative error in % of clustered matrix approximation
nA = norm(A,'fro'); relerrCmapp = sqrt( nA^2 - norm(SS, 'fro')^2 )/nA*100; relerrCmappR = sqrt( nA^2 - norm(SSr,'fro')^2 )/nA*100; % Compute memory consumption memCmapp = memUsageCmapp(U,SS,V,'non-sym'); memCmappR = memUsageCmapp(Ur,SSr,Vr,'non-sym'); % Get rank k for a regular matrix approximation with = or > memory usage [m,n] = size(A); [memRmapp,kreg] = memUsageRmapp(m,n,memCmapp,'sym'); [Ur,Sr,Vr] = svds(A,kreg); relerrRmapp = sqrt( nA^2 - norm(Sr,'fro')^2 )/nA*100; % fpns = floating point numbers disp('-------------------------------------------------------------------') fprintf('Clustered matrix approximation: relErr = %10.2f %% \n', relerrCmapp) fprintf('Clustered matrix approximation: relErr = %10.2f %% (randomized) \n', relerrCmappR) fprintf('Regular matrix approximation: relErr = %10.2f %% \n', relerrRmapp) disp('-------------------------------------------------------------------') fprintf('Clustered matrix approximation: memUsage = %10.0f fpns \n', memCmapp) fprintf('Clustered matrix approximation: memUsage = %10.0f fpns (randomized) \n', memCmappR) fprintf('Regular matrix approximation: memUsage = %10.0f fpns \n', memRmapp) disp('-------------------------------------------------------------------')
------------------------------------------------------------------- Clustered matrix approximation: relErr = 94.85 % Clustered matrix approximation: relErr = 95.08 % (randomized) Regular matrix approximation: relErr = 97.51 % ------------------------------------------------------------------- Clustered matrix approximation: memUsage = 12065020 fpns Clustered matrix approximation: memUsage = 13481006 fpns (randomized) Regular matrix approximation: memUsage = 12109952 fpns -------------------------------------------------------------------